Afni Builds For Mac
Development of CHIRP is an all-volunteer effort and is offered as open-source software, free of charge. If you like CHIRP, please consider contributing a small donation to help support the costs of development and hardware:CHIRP downloadsCHIRP is distributed as a series of automatically-generated builds. Any time we make a change to CHIRP, a build is created for it the next day. Thus, CHIRP is versioned by the date on which it was created, which makes it easy to determine if you have an older build. We don't put experimental things into CHIRP before they are ready, except where specifically called out with a warning. Thus, you do not need to worry about finding a stable version to run. You should always be on the latest build available.Upgrading: You do NOT need to uninstall an existing version of CHIRP before installing a newer one.
What does AFNI stand for? List of 13 AFNI definitions. Updated April 2020. Top AFNI abbreviation meaning: Adjusted Family Net Income.
Just install the new one and it will replace the existing copy!You can find a complete test report of the current build and a matrix of supported models and features Windows Users. CHIRP runs on Windows 2000, XP, Vista, 7, 8, and 10.
Older versions of Windows are not supported. Most users will want to download the installer.exe file, which installs CHIRP like a normal application.
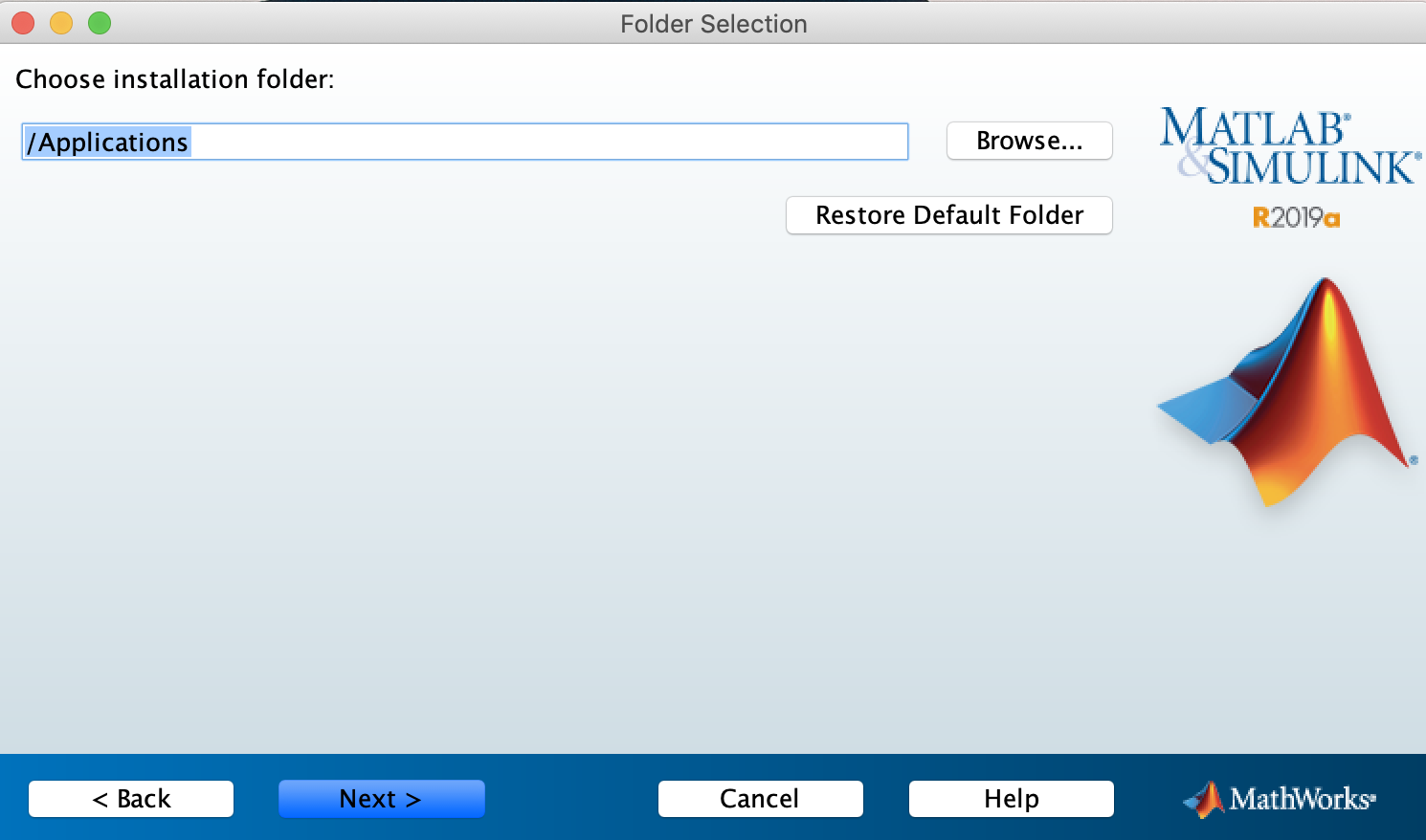
The win32.zip file is for advanced users wishing to run CHIRP without installingMacOS Users. New Mac OS X must install the runtime once before running CHIRP. After it is installed, the runtime does not need to be downloaded or installed each time. Download the for Mac OSX.
OS X support is limited to Intel architecture. PowerPC is not supported. Chirp.app is unsigned. With default security settings, you will need a special procedure to run it:.users can install Chirp without the KK7DS runtime by running brew install tdsmith/ham/chirp and then running chirp from the terminal.However, running CHIRP using Homebrew is obsolete. MacOS Unified Application build is now the recommended way.More useful tidbits can be found at.
Ubuntu Linux UsersIf you are using Ubuntu linux (or a compatible variant such as Mint) you should install and use the PPA like this: sudo apt-add-repository ppa:dansmith/chirp-snapshotssudo apt-get updatesudo apt-get install chirp-dailyAfter installing the build from the PPA, new updates will be included in your normal system software updates. Also see the page for additional steps required to gain access to your serial port. Other Linux UsersIf you don't run a distro for which we have packages, you can run CHIRP right from the tarball available through the link above. Most modern distributions should have almost everything required to run chirp. Make sure you have python-serial and python-libxml2 packages installed.
For more information about using CHIRP under Linux, see the page Translators and DevelopersThere is extensive documentation about the development and contribution process located at Old VersionsYou can access older versions of chirp here:.
Sample AFNI session.Robert W. Cox WebsiteAnalysis of Functional NeuroImages ( AFNI) is an environment for processing and displaying data—a technique for mapping human brain activity.AFNI is an agglomeration of programs that can be used interactively or flexibly assembled for using.
The term AFNI refers both to the entire suite and to a particular interactive program often used for visualization. AFNI is actively developed by the NIMH Scientific and Statistical Computing Core and its capabilities are continually expanding.AFNI runs under many operating systems that provide and libraries, including,. Precompiled binaries are available for some platforms. AFNI is available for research use under the. AFNI now comprises over 300,000 lines of, and a skilled C programmer can add interactive and batch functions to AFNI with relative ease. Contents.History and development AFNI was originally developed at the beginning in 1994, largely by Robert W.
Cox brought development to the NIH in 2001 and development continues at the NIMH Scientific and Statistical Computing Core. In a 1995 paper describing the rationale for development of the software, Cox wrote of fMRI data: 'The volume of data gathered is very large, and it is essential that easy-to-use tools for visualization and analysis of 3D activation maps be available for neuroscience investigators.' Since then, AFNI has become one of the more commonly used analysis tools in fMRI research, alongside and.Although AFNI initially required extensive shell scripting to execute tasks, pre-made batch scripts and improvements to the have since made it possible to generate analyses with less user scripting. Features Visualization One of AFNI's initial offerings improved the approach to transforming scans of individual brains onto a shared standardized space.
Since each person's individual brain is unique in size and shape, comparing across a number of brains requires warping (rotating, scaling, etc.) individual brains into a standard shape. Unfortunately, functional MRI data at the time of AFNI's development was too low resolution for effective transformations. Instead, researchers use the higher resolution anatomical brain scans, often acquired at the beginning of an imaging session.AFNI allows researchers to overlay a functional image to the anatomical, providing tools for aligning the two into the same space. Processes engaged to warp an individual anatomical scan to standard space are then applied also to the functional scan, improving the transformation process.Another feature available in AFNI is the SUMA tool, developed by Ziad Saad. This tool allows users to project the 2D data onto a 3D cortical surface map. In this way researchers can view activation patterns while more easily taking into account physical cortical features like gyri.
Image Pre-processing 'afniproc.py' is a pre-made script that will run fMRI data from a single subject through a series of pre-processing steps, starting with the raw data. The default settings will perform the following pre-processing steps and finish with a basic regression analysis:.
Slice timing: Each 3D brain image is composed of multiple 2D images, 'slices'. Although acquired at approximately the same time, up to several seconds could separate the first slice acquired from the last. Through interpolation, the slices are aligned to the same time point. Generally, any introduced noise from interpolation errors is thought to be outweighed by improvements in signal. Motion correction: Head movements can create sources of error in the analysis. Each 3D acquisition in a scan is collected on a 3D grid, with each small cube of grid space, ', representing a single image intensity value. Ideally, voxels will always represent the same part of the brain in each acquisition, rather than vary from one 3D image to the next.
To correct small motion artifacts, AFNI's motion correction tool employs a linear least squares algorithm that attempts to align each 3D image acquired to the first image acquired in the scan. Smoothing: To account for random noise in the image, a smoothing kernel is applied.
While smoothing can increase the signal-to-noise ratio of the image, it reduces image resolution. Mask: Removes any non-brain areas, such as skull, from the fMRI image. Scale: Scale each voxel so that changes in intensity represent percentage of signal change over the course of the scan. The default sets the mean of each voxel equal to 100.See also. Questions and Answers in MRI. Retrieved 2018-05-14. Cox, Robert W.
'AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages'. Computers and Biomedical Research. 29 (3): 162–173. Murnane, Kevin. Retrieved 2018-05-14. Jahn, Andrew (2012-12-28).
Andy's Brain Blog. Retrieved 2018-05-21. Cox, Robert W. 'AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages'.
Computers and Biomedical Research. 29 (3): 162–173. 1941 hong kong on fire. Jahn, Andrew (2012-03-26). Andy's Brain Blog.
Retrieved 2018-05-14. Retrieved 2018-05-21. Retrieved 2018-05-21. Retrieved 2018-05-21.
Retrieved 2018-05-21. Retrieved 2018-05-21. Retrieved 2018-05-21.External links.